ASPL Guide to Operations v 1.00
© 2024 by SetSphere.COM
9. 7g%& aligns two sequences based on their subgroups and the elements intersection
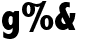
g%& a1 a2 | a1 a2 a3 where a1 a2 | a1 a2 a3 are set variables
g%&`mtm~ a1 a2 | a1 a2 a3
g%&/mtm~ a1 a2 | a1 a2 a3
g%&
When specified by itself then the operation is performed on the object on top of the stack and the one that is one below it
seqop seqoid 23 notcommu ins:H_H_H text:alignment
Use the operator g%& to align two set variables according to their subgroups and elements, and display their alignment on the terminal. It is assumed that each set variable has one group label, otherwise the first group (in the set variable) is explicitly implied. If you specify two set variables that have the same named group label, then you need to labinate the operator (see example below). If the operator g%& is followed by three set variables then the alignment is performed on the datasets resulted by concatenating the third variable to the first and to the second. Hence this is similar to performing an alignment after joining with an "and" operation the first variable and the third variable, and the second variable and the third variable. You can change the sequence alignment algorithm selectively. The sequence alignment can be selected: the command sequencing lcs selects the algorithm for the longest common sequence, and the command sequencing ssa selects the algorithm for the sequence symmetry analysis.
g%& EXAMPLEIn this example we will compare two directories by displaying their sequence alignment.
We will start ASPL by loading the WS1 workspace, then we will issue the command g%& to display the alignment of the groups (that is subgroups and elements) represented by the two set variables a1 and a2. Note that the sample worskpace WS1 contain datasets representing UNIX directory contents, in which case the subgroups and elements of a set variable are equivalent to the subdirectories and files in the datasets. Refer to sample workspace WS1 for more information.
g%& OPERATION# aspl WS1
(start ASPL loading the sample workspace WS1)
① aspl>
g%& a1 a2
(show the alignment of the subgroups and elements between a1 and a2)
We expect to have perfect alignment if two set variables are similar. As an example we align a1 and a copy of itself.
② aspl>
a11 = a1
(a11 is assigned to a1 so a11 and a1 are similar)
③ aspl>
v
(display the symbol table, notice that a1 and a11 have the same label under the Lvr column)
④ aspl>
g%& a11 a1
(align the operator based on their elements but nothing is displayed)
⑤ aspl>
g%&, a11 a1
(labinate the operator g%& to show the perfect alignment between a1 and a11 )
⑥ aspl>
,g%& a11 a1
(the labinate comma can also precede the operator)
● Tickable Predicates for g%&
This g%& is tickable with predicates that are shown in the following table:
a yellow note
g%&`_ aligns two sequences based on their subgroups and the elements intersection g%&`c= aligns two sequences based on their subgroups and the elements intersection such that they have the same checksum g%&`c~ aligns two sequences based on their subgroups and the elements intersection such that they have different checksums g%&`c~~ aligns two sequences based on their subgroups and the elements intersection such that they have all over different checksums g%&`h= aligns two sequences based on their subgroups and the elements intersection such that they have the same entropy g%&`h~ aligns two sequences based on their subgroups and the elements intersection such that they have different entropies g%&`h~~ aligns two sequences based on their subgroups and the elements intersection such that they have all over different entropies g%&`mtm= aligns two sequences based on their subgroups and the elements intersection such that they have the same make time g%&`mtm~ aligns two sequences based on their subgroups and the elements intersection such that they have different make times g%&`mtm~~ aligns two sequences based on their subgroups and the elements intersection such that they have all over different make times g%&`mtm< aligns two sequences based on their subgroups and the elements intersection such that they have a prior make time g%&`mtm<= aligns two sequences based on their subgroups and the elements intersection such that they have prior or same make time g%&`mtm> aligns two sequences based on their subgroups and the elements intersection such that they have more recent make time g%&`mtm>= aligns two sequences based on their subgroups and the elements intersection such that they have more recent or same make time g%&`ks= aligns two sequences based on their subgroups and the elements intersection such that they have the same ksum g%&`ks~ aligns two sequences based on their subgroups and the elements intersection such that they have different ksums g%&`ks~~ aligns two sequences based on their subgroups and the elements intersection such that they have all over different ksums